


(4 November 2025)
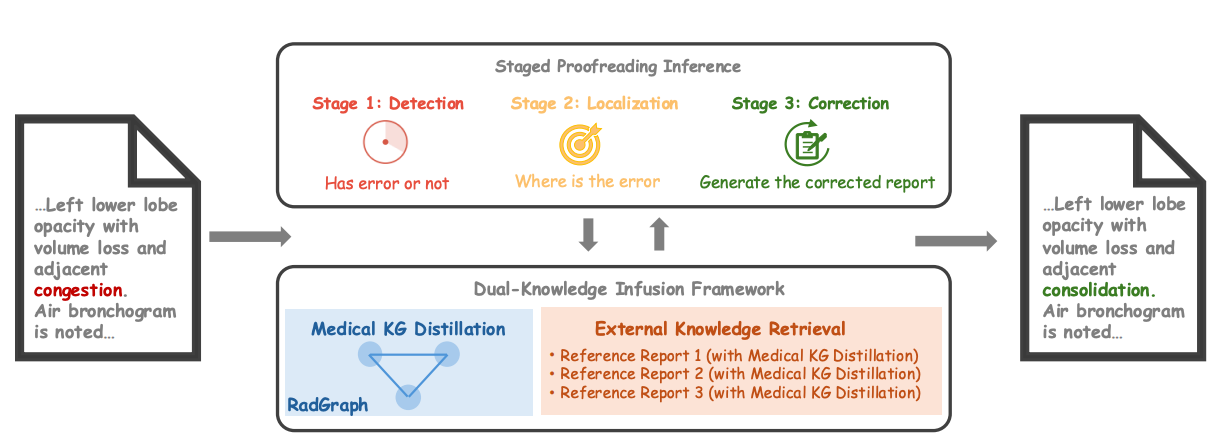 Paper accepted by AAAI 2026 Artificial Intelligence for Social Impact Track – Error Correction in Radiology Reports - A Knowledge Distillation-Based Multi-Stage Framework at https://arxiv.org/abs/2406.15045v3
Paper accepted by AAAI 2026 Artificial Intelligence for Social Impact Track – Error Correction in Radiology Reports - A Knowledge Distillation-Based Multi-Stage Framework at https://arxiv.org/abs/2406.15045v3
The increasing complexity and workload of clinical radiology leads to inevitable oversights and mistakes in their use as diagnostic tools, causing delayed treatments and sometimes life-threatening harms to patients. While large language models (LLMs) have shown remarkable progress in many tasks, their utilities in detecting and correcting errors in radiology reporting are limited. We present a novel dual-knowledge infusion framework that enhances LLMs’ capability for radiology report proofreading through systematic integration of medical expertise. Specifically, our knowledge infusion combines medical knowledge graph distillation (MKGD) with external knowledge retrieval (EXKR), enabling an effective automated approach in tackling mistakes in radiology reporting. By decomposing the complex proofreading task into three specialized stages of detection, localization, and correction, our method mirrors the systematic review process employed by expert radiologists, ensuring both precision and clinical interpretability. The dual-knowledge framework captures intricate medical relationships through structured graph representations while leveraging curated clinical expertise from reference reports. To perform a robust, clinically relevant evaluation, we constructed a comprehensive benchmark using real-world radiology reports with error patterns derived from real-world scenarios, including speech recognition confusions, terminology ambiguities, and template-related inconsistencies, all validated by practicing radiologists. Extensive evaluations across multiple LLM architectures demonstrate substantial improvements of our approach - up to 31.56% increase in error detection accuracy and 37.4% reduction in processing time. Human evaluation by radiologists confirms superior clinical relevance and factual consistency compared to existing approaches. Our framework addresses critical needs in clinical practice by enhancing report quality while reducing radiologist burden, particularly benefiting resource-constrained healthcare environments.
Read the preprint version at https://arxiv.org/abs/2406.15045v3.
(31 October 2025)
 A systematic review published on Communications Medicine - Digital health technology use in clinical trials of rare diseases - a systematic review at https://www.nature.com/articles/s43856-025-01137-6
A systematic review published on Communications Medicine - Digital health technology use in clinical trials of rare diseases - a systematic review at https://www.nature.com/articles/s43856-025-01137-6
Hundreds of millions of people around the world live with rare diseases, but most of these conditions still don’t have effective treatments. Clinical trials are vital for new therapies, but rare disease trials are especially challenging due to small, dispersed patient populations and difficulties with long-term participation. This study looked at how digital health tools—like wearable devices, mobile apps, and healthcare platforms—are being used to improve rare disease clinical trials. We analysed 262 trials across ten rare diseases and found that digital health technologies were increasingly used for remote monitoring, digital treatment, and long-term care. By making it easier for patients to participate in trials—no matter where they live—digital technologies can help speed up the development of much-needed treatments and help make research more inclusive and equitable by reaching patients who would otherwise be left out.
Read the full article at https://www.nature.com/articles/s43856-025-01137-6.
(21 October 2025)
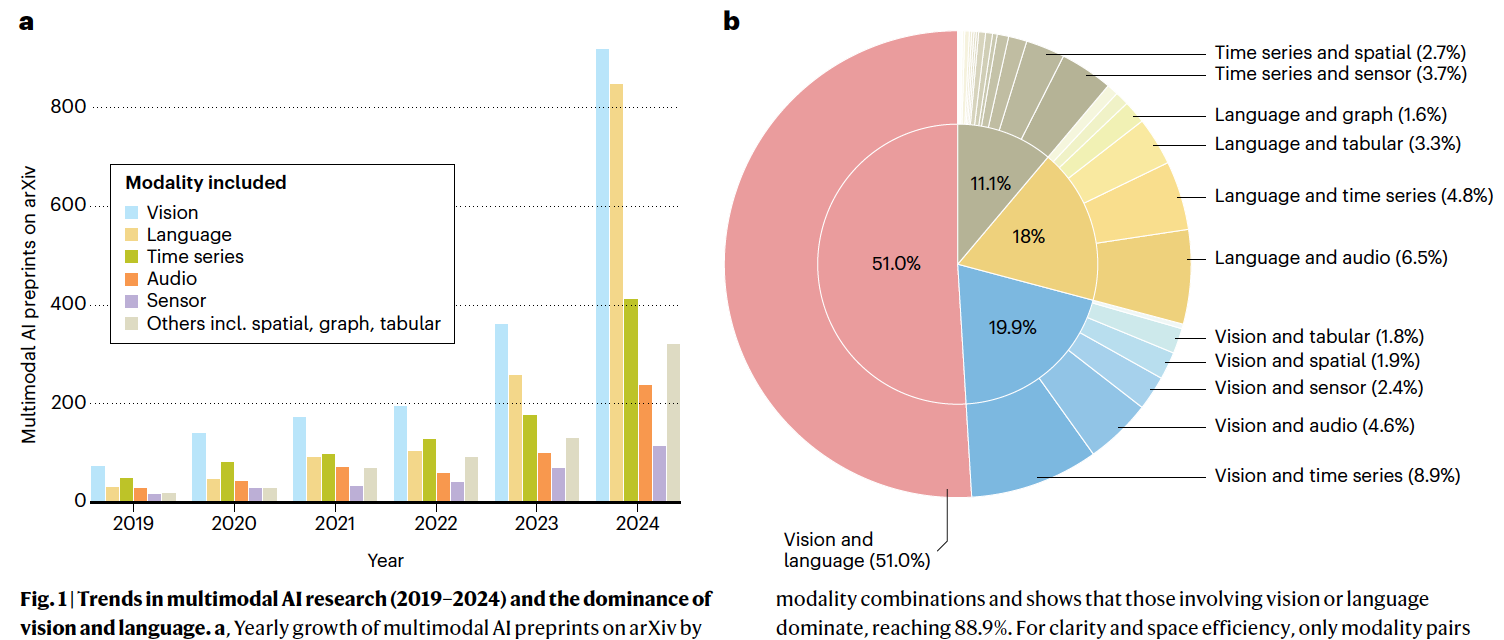 A collaborative, perspective paper published on Nature Machine Intelligence - Towards deployment-centric multimodal AI beyond vision and languag at https://www.nature.com/articles/s42256-025-01116-5
A collaborative, perspective paper published on Nature Machine Intelligence - Towards deployment-centric multimodal AI beyond vision and languag at https://www.nature.com/articles/s42256-025-01116-5
Multimodal artificial intelligence (AI) integrates diverse types of data via machine learning to improve understanding, prediction and decision-making across disciplines such as healthcare, science and engineering. However, most multimodal AI advances focus on models for vision and language data, and their deployability remains a key challenge. We advocate a deployment-centric workflow that incorporates deployment constraints early on to reduce the likelihood of undeployable solutions, complementing data-centric and model-centric approaches. We also emphasize deeper integration across multiple levels of multimodality through stakeholder engagement and interdisciplinary collaboration to broaden the research scope beyond vision and language. To facilitate this approach, we identify common multimodal-AI-specific challenges shared across disciplines and examine three real-world use cases - pandemic response, self-driving car design and climate change adaptation, drawing expertise from healthcare, social science, engineering, science, sustainability and finance. By fostering interdisciplinary dialogue and open research practices, our community can accelerate deployment-centric development for broad societal impact.
Read the full article at https://www.nature.com/articles/s42256-025-01116-5.
(19 September 2025)
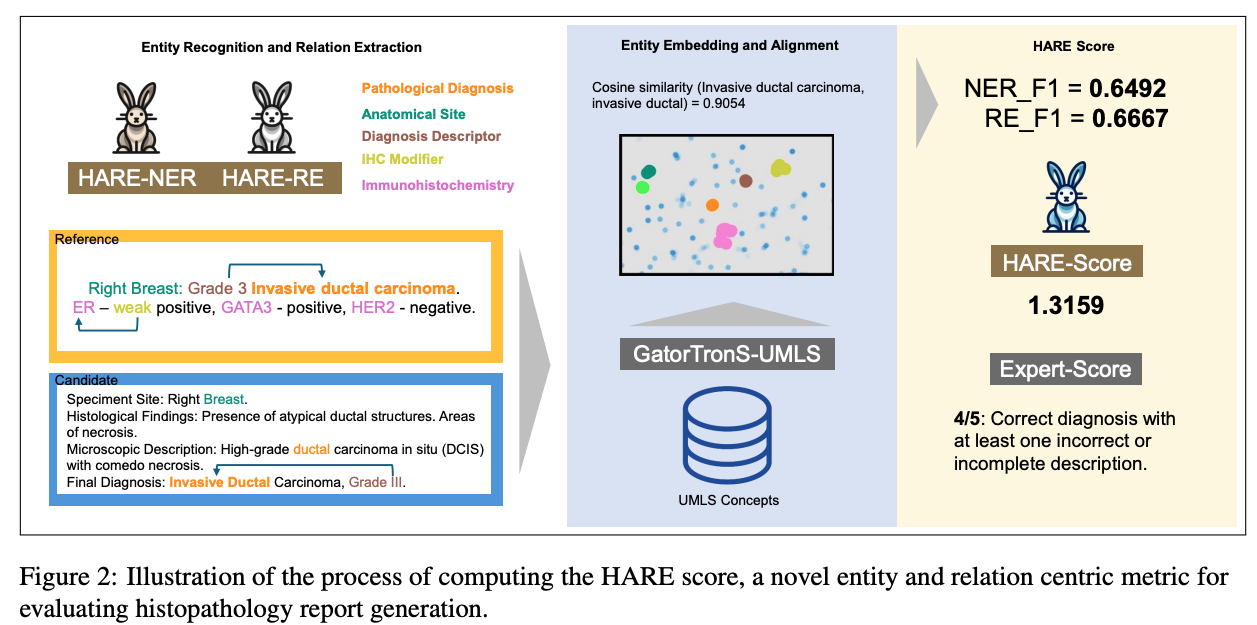 Paper accepted by EMNLP 2025 findings – HARE- an entity and relation centric evaluation framework for histopathology reports – at https://arxiv.org/abs/2507.09097
Paper accepted by EMNLP 2025 findings – HARE- an entity and relation centric evaluation framework for histopathology reports – at https://arxiv.org/abs/2507.09097
Large Vision-Language Models (LVLMs) have demonstrated promising performance in chest X-ray (CXR) analysis. To enhance human-computer interaction, several studies have incorporated radiologists’ eye gaze, typically through heatmaps or textual prompts. However, these methods often overlook the sequential order of eye movements, which could provide valuable insights by highlighting both the areas of interest and the order in which they are examined. In this work, we propose a novel approach called RadEyeVideo that integrates radiologists’ eye-fixation data as a video sequence, capturing both the temporal and spatial dynamics of their gaze. We evaluate this method in CXR report generation and disease diagnosis using three general-domain, open-source LVLMs with video input capabilities. When prompted with eye-gaze videos, model performance improves by up to 24.6% in the report generation task and on average 15.2% for both tasks using scaled evaluation metrics. Notably, RadEyeVideo enhanced an open-domain LVLM model, LLaVA-OneVision, to surpass task-specific medical LVLMs such as MAIRA-2 and CheXagent, trained on large Chest X-ray data. This work highlights that domain expert’s knowledge (eye-gaze information in this case), when effectively integrated with LVLMs, can significantly enhance general-domain models’ capabilities in clinical tasks. RadEyeVideo is a step toward a scalable human-centered approach of utilizing LVLMs in medical image analytics.
Read the full paper at https://arxiv.org/abs/2507.09097.
(8 August 2025)
 Tokenizer work published - Infusing clinical knowledge into language models by subword optimisation and embedding initialisation - now with Computers in Biology and Medicine.
Tokenizer work published - Infusing clinical knowledge into language models by subword optimisation and embedding initialisation - now with Computers in Biology and Medicine.
This study introduces a novel knowledge enhanced tokenisation mechanism, K-Tokeniser, for clinical text processing. Technically, at The study proposes a novel tokenisation method utilising global representations of tokens based on domain-specific concepts (e.g., drugs, diseases) from ontologies like UMLS or task-specific corpora. At training or inference, word and sentence-level optimisation is used to select the optimal token representations. It proposes an embedding initialisation approach for new tokens, eliminating the need for pre-training the language models. The Model built using K-Tokeniser achieves a notable 13% increase in Micro F1 score for automated clinical coding. It requires only 50% of training data for concept extraction and less than 20% for automated coding to outperform the baseline clinicalBERT model.
Read it at 10.1016/j.compbiomed.2025.110747.
(28 May 2025)
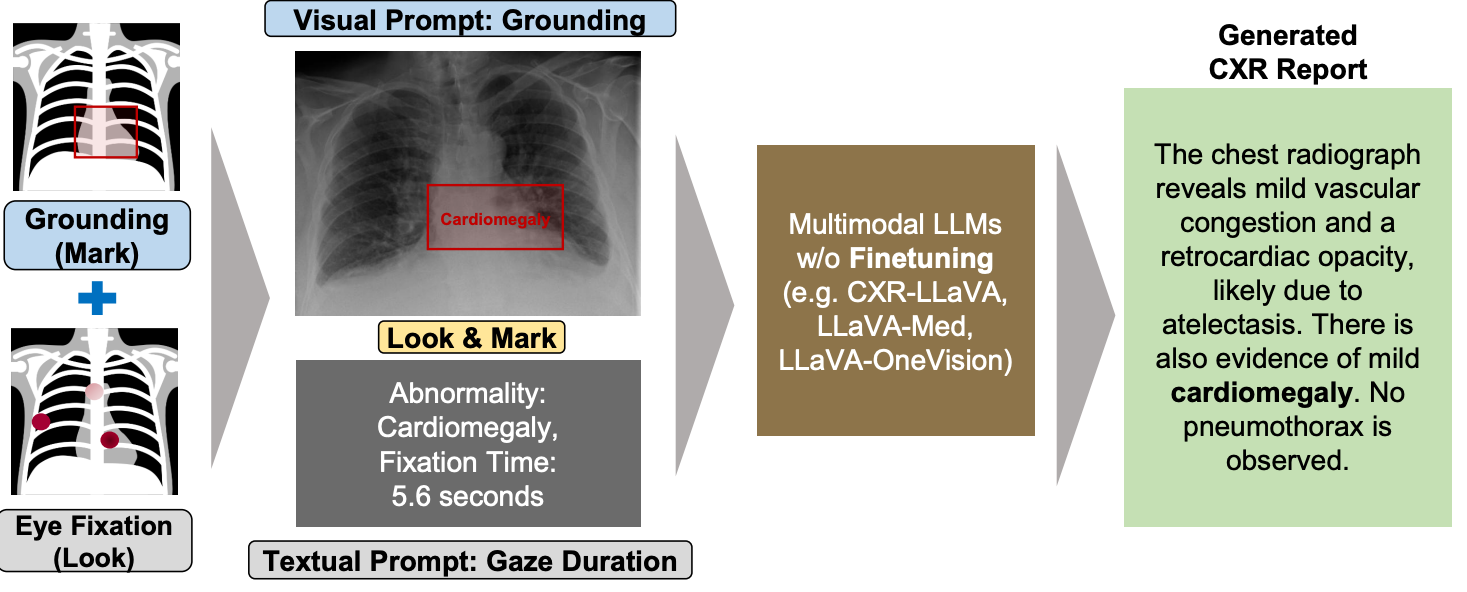 Paper accepted by ACL 2025 findings – Look & Mark - Leveraging Radiologist Eye Fixations and Bounding boxes in Multimodal Large Language Models for Chest X-ray Report Generation – at https://arxiv.org/abs/2505.22222
Paper accepted by ACL 2025 findings – Look & Mark - Leveraging Radiologist Eye Fixations and Bounding boxes in Multimodal Large Language Models for Chest X-ray Report Generation – at https://arxiv.org/abs/2505.22222
This study introduces Look & Mark (L&M), a novel approach to radiology report generation that integrates radiologist fixation cues (Look) with bounding box annotations (Mark) to guide multimodal Large Language Models (LLMs). By combining these complementary grounding strategies, L&M significantly improves the clinical relevance of generated reports, reduces hallucinations, and enhances model alignment with real-world diagnostic workflows. Importantly, L&M achieves these gains without requiring extensive fine-tuning, leveraging in-context learning to adapt both general-purpose and domain-specific models alike.
Our experiments demonstrate that L&M significantly boosts performance across both lexical and clinical evaluation metrics, with the largest gains observed in clinical metrics such as RaTEScore and RadGraph-XL. For instance, CXR-LLaVA achieved a 1.2% improvement in overall metrics (A.AVG) compared to baseline prompting, while LLaVA-Med demonstrated a remarkable 9.2% boost. General-purpose models also benefited significantly, with LLaVA-OV achieving an 87.3% clinical average (C.AVG), the highest among all tested models, even surpassing domain-specific models trained explicitly for chest X-ray report generation.
Read the full paper at https://arxiv.org/abs/2505.22222.
(28 May 2025)
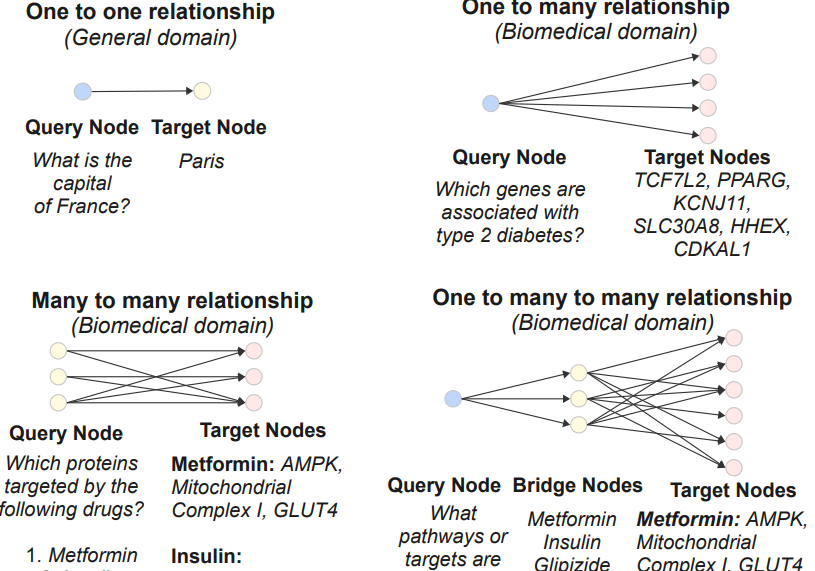 Paper accepted by ACL 2025 findings – BioHopR - A Benchmark for Multi-Hop, Multi-Answer Reasoning in Biomedicine – at https://arxiv.org/abs/2505.22240
Paper accepted by ACL 2025 findings – BioHopR - A Benchmark for Multi-Hop, Multi-Answer Reasoning in Biomedicine – at https://arxiv.org/abs/2505.22240
This paper introduces BioHopR, a benchmark for evaluating multi-hop, multi-answer reasoning in the biomedical domain. Built on the PrimeKG knowledge graph, BioHopR captures the complexity of real-world biomedical queries through one-tomany and many-to-many relationships, rigorously assessing reasoning over 1-hop and 2-hop tasks. Evaluation results highlight that O3-mini, a proprietary model with a reasoning step, outperforms open-source models including biomedical models like HuatuoGPT-o1. Across all models, the performance drop from 1-hop to 2-hop tasks underscores the difficulty of aligning intermediate reasoning steps, especially in bridging entities.
Read the full paper at https://arxiv.org/abs/2505.22240.
(12 May 2025)
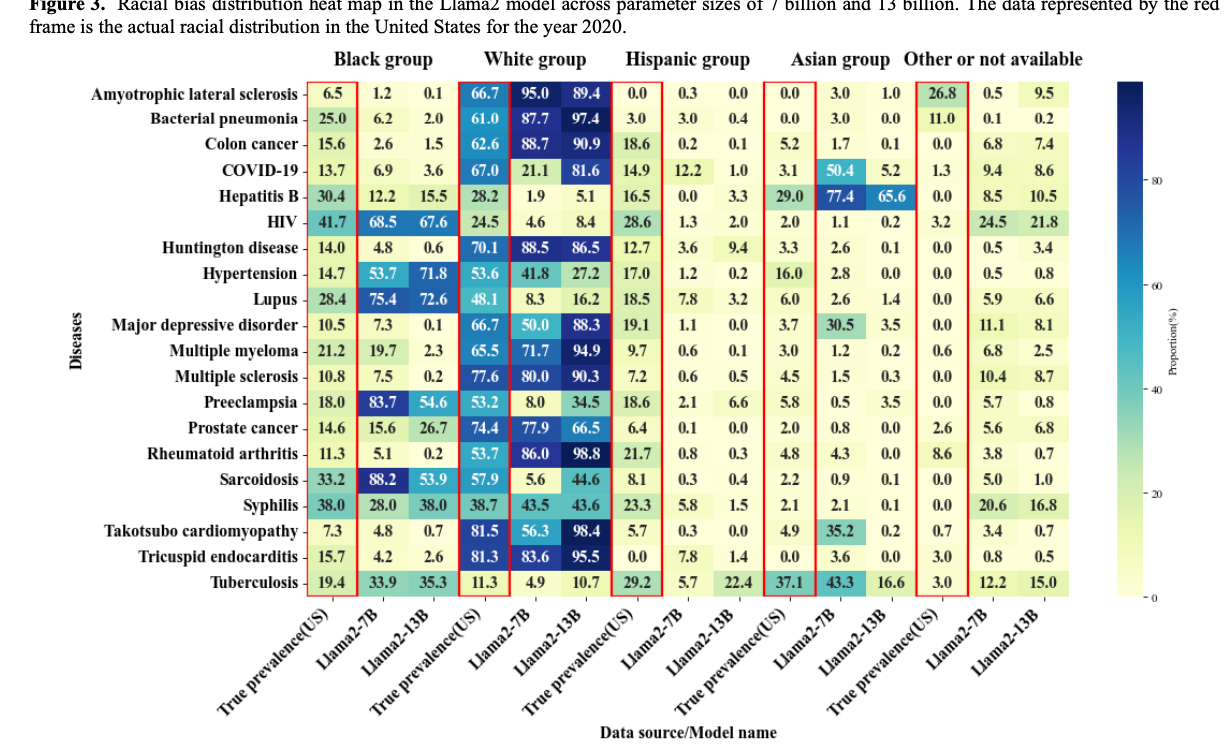 Paper accepted by Journal of Medical Internet Research – Evaluation and Bias Analysis of Large Language Models in Generating Synthetic Electronic Health Records - Comparative Study – at https://doi.org/10.2196/65317
Paper accepted by Journal of Medical Internet Research – Evaluation and Bias Analysis of Large Language Models in Generating Synthetic Electronic Health Records - Comparative Study – at https://doi.org/10.2196/65317
Larger models, such as Yi-34B, Qwen-14B, and Llama 2 to 13 B, showed improved performance in generating more comprehensive EHRs, as reflected in higher EPS values. However, this increased performance was accompanied by a notable escalation in both gender and racial biases, highlighting a performance-bias trade-off. The study identified 4 key findings as follows (1) as model size increased, EHR generation improved, but demographic biases also became more pronounced; (2) biases were observed across all models, not just the larger ones; (3) gender bias closely aligned with real-world disease prevalence, while racial bias was evident in only a subset of diseases; and (4) racial biases varied, with some diseases showing overrepresentation of White or Black populations and underrepresentation of Hispanic and Asian groups. These findings underline the need for effective bias mitigation strategies and the development of benchmarks to ensure fairness in artificial intelligence applications for health care.
Read the full paper at https://doi.org/10.2196/65317.
(18 April 2025)
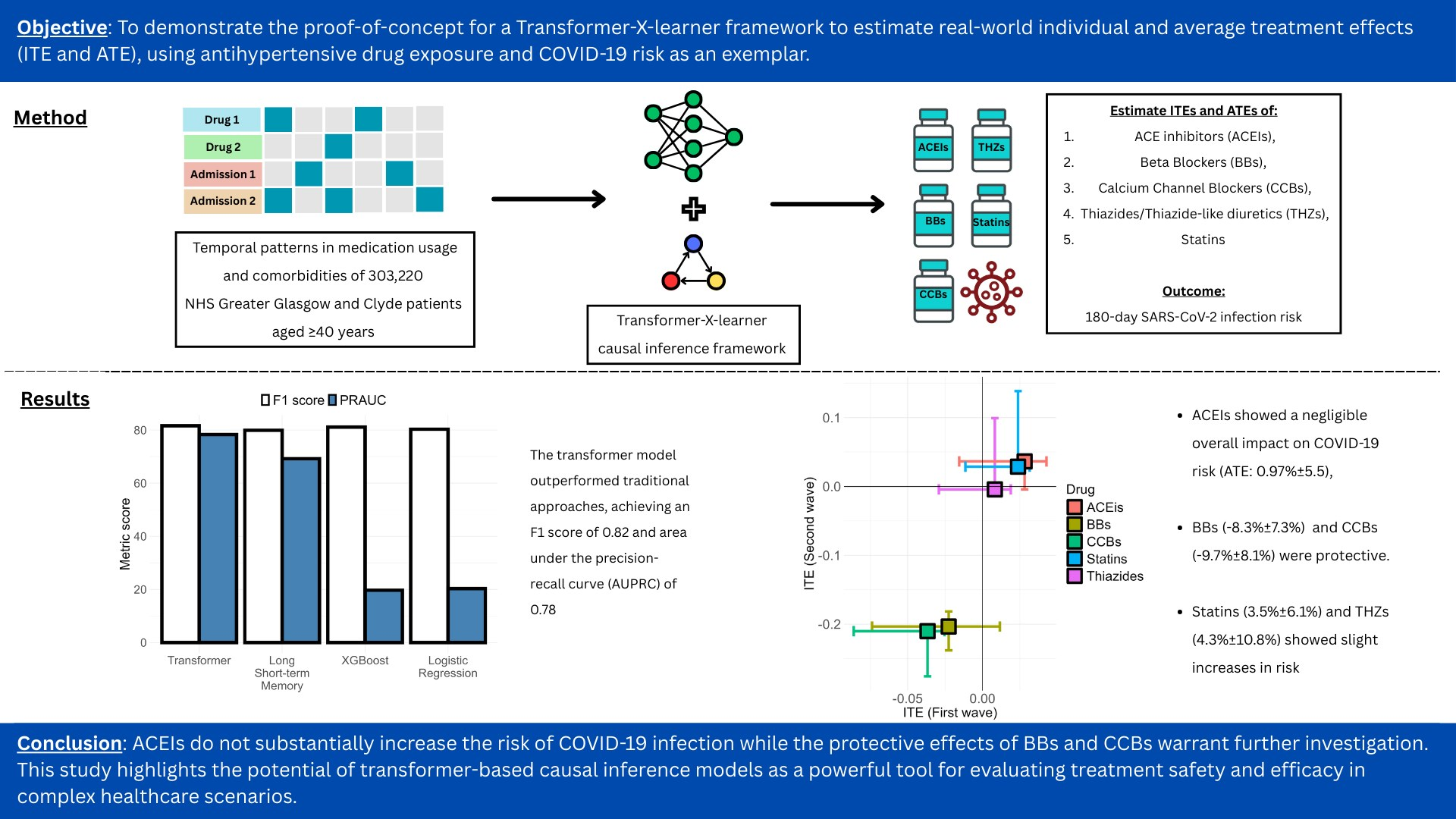 Published in American Journal of Hypertension – A Transformer-Based Framework for Counterfactual Estimation of Antihypertensive Treatment Effect on COVID-19 Infection Risk - A Proof-of-Concept Study – at https://doi.org/10.1093/ajh/hpaf055
Published in American Journal of Hypertension – A Transformer-Based Framework for Counterfactual Estimation of Antihypertensive Treatment Effect on COVID-19 Infection Risk - A Proof-of-Concept Study – at https://doi.org/10.1093/ajh/hpaf055
A new study in the American Journal of Hypertension investigates the relationship between antihypertensive medications and COVID-19 infection risk. The research employed a transformer-based framework to analyze real-world data from over 300,000 patients.
Key findings indicate that while ACE inhibitors showed a negligible effect on COVID-19 risk, beta-blockers and calcium channel blockers were associated with a protective effect. Statins and thiazides showed a slight increase in risk.
This study demonstrates the potential of advanced causal inference models in evaluating treatment outcomes in complex healthcare scenarios and offers important insights for clinical consideration.
Read the full paper at https://doi.org/10.1093/ajh/hpaf055.
(8 April 2025)
 Paper published in Journal of Imaging Informatics in Medicine – How Do Radiologists Currently Monitor AI in Radiology and What Challenges Do They Face? – at DOI:10.1007/s10278-025-01493-8
Paper published in Journal of Imaging Informatics in Medicine – How Do Radiologists Currently Monitor AI in Radiology and What Challenges Do They Face? – at DOI:10.1007/s10278-025-01493-8
As AI tools become more common in radiology, monitoring their performance is increasingly important—but still underdeveloped. A recent qualitative study interviewed 16 radiologists across Europe and the U.S., revealing that many AI systems are still in early validation phases. Current monitoring typically involves manual, retrospective comparisons to radiology reports—effective, but labor-intensive.
Key barriers include a lack of standardized monitoring guidelines, limited technological tools, and constrained resources. The study recommends mixed-method monitoring, dedicated governance teams, and long-term resource planning.
This research highlights the need for clearer frameworks and investment to ensure AI improves clinical workflows.
Read it at DOI:10.1007/s10278-025-01493-8.
(4 April 2025)
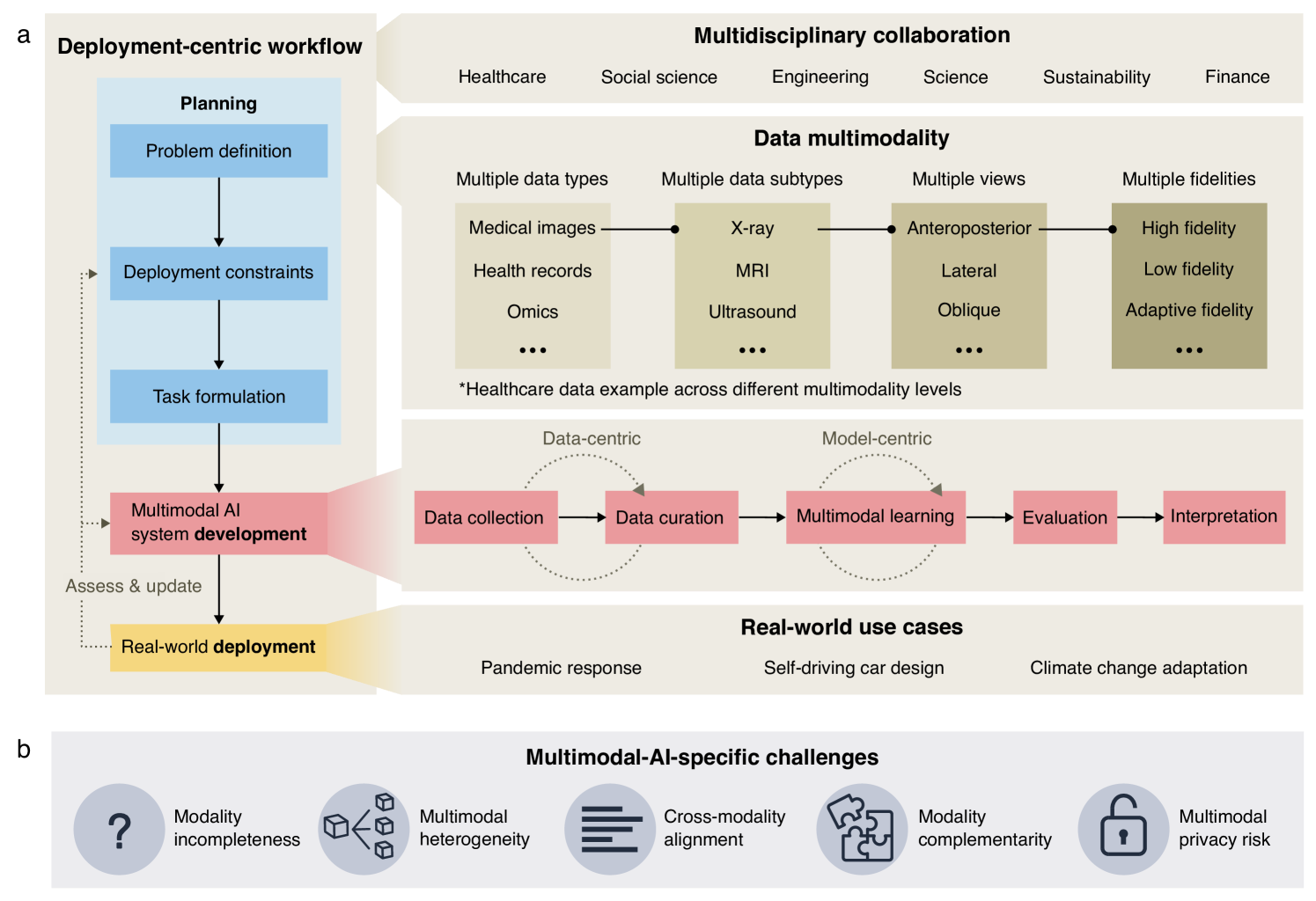 Preprint published on arXiv – Towards Deployment-Centric Multimodal AI Beyond Vision and Language – at arXiv:2504.03603v1
Preprint published on arXiv – Towards Deployment-Centric Multimodal AI Beyond Vision and Language – at arXiv:2504.03603v1
This work introduces a deployment-centric workflow for multimodal AI, emphasizing real-world applicability beyond just vision and language models. While multimodal systems hold immense potential in areas like healthcare and engineering, deployment challenges are often an afterthought. This paper pushes for a proactive approach—integrating data readiness, model robustness, and system integration into early development.
By shifting focus from just performance benchmarks to deployment feasibility, this research bridges the gap between prototypes and practical implementation.
It’s a compelling case for aligning AI innovation with real-world impact.
Read it at arXiv:2504.03603v1.
(1 April 2025)
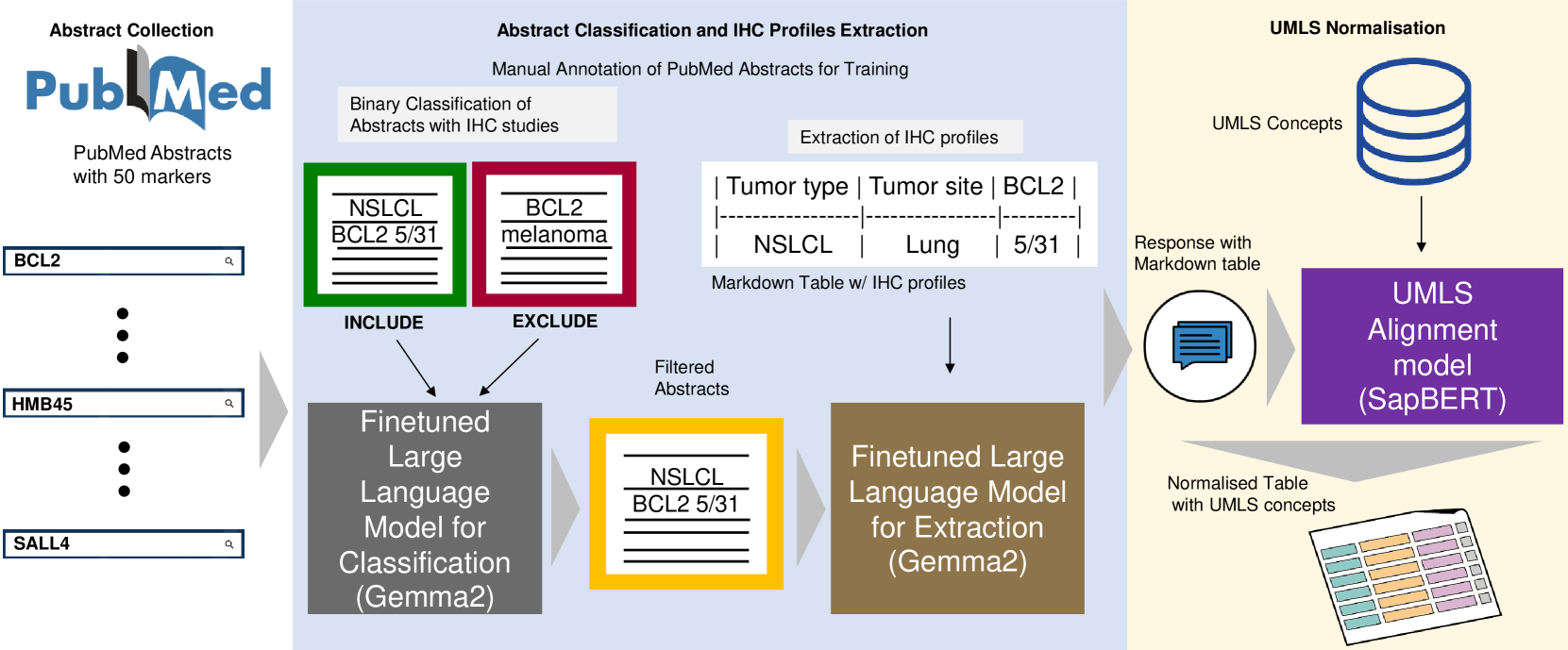 Preprint published on arXiv – IHC-LLMiner- Automated extraction of tumour immunohistochemical profiles from PubMed abstracts using large language models – at arXiv:2504.00748v1
Preprint published on arXiv – IHC-LLMiner- Automated extraction of tumour immunohistochemical profiles from PubMed abstracts using large language models – at arXiv:2504.00748v1
This study introduces IHC-LLMiner, an automated pipeline that extracts immunohistochemical (IHC) tumour profiles from PubMed abstracts using large language models (LLMs). The pipeline performs two tasks - classifying abstracts for relevance and extracting IHC-tumour profiles from relevant abstracts.
The fine-tuned Gemma-2 model achieved 91.5% accuracy in classification and 63.3% correctness in profile extraction, outperforming GPT4-O in both accuracy and speed. Extracted profiles were normalized to UMLS concepts, facilitating consistent analysis.
IHC-LLMiner demonstrates potential for large-scale IHC data mining, enhancing accessibility and utility for research and clinical applications.
Read it at arXiv:2504.00748v1.